Dr. Yu-Chiao (Chris) Chiu, Assistant Professor of Medicine in the Hematology/Oncology department at Hillman cancer center published a paper in a Cell Press journal “Patterns” entitled, “shinyDeepDR: A user-friendly R Shiny app for predicting anti-cancer drug response using deep learning“. Dr. Paul Monga was a coauthor on the article and Dr Chiu was funded in part by a PLRC P&F grant.
-
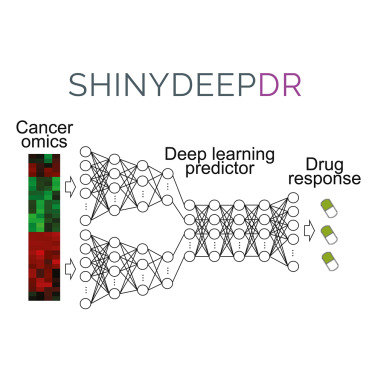
shinyDeepDR is a web tool for predicting responses to 265 anti-cancer compounds
- It is applicable for researching both cancer cell lines and tumors
- Its interactive web interface facilitates result interpretation and investigation
- It identifies promising targets for an “undruggable” mutation in liver cancer
The Web tool is available online at: https://shiny.crc.pitt.edu/shinydeepdr/
Li-Ju Wang, Michael Ning, Tapsya Nayak, Michael J. Kasper, Satdarshan P. Monga, Yufei Huang, Yidong Chen, Yu-Chiao Chiu, shinyDeepDR: A user-friendly R Shiny app for predicting anti-cancer drug response using deep learning,Patterns,2024,100894,ISSN 2666 3899, https://doi.org/10.1016/j.patter.2023.100894.
The bigger picture
Understanding how different genomic attributes affect drug responses in cancer is crucial for personalized oncology. Deep learning, an advanced computational method, has demonstrated significant potential in identifying and predicting these intricate interactions. One such example is the DeepDR model, which predicts how cancer cells respond to drugs. However, not all researchers have the computational resources and programming expertise to leverage this potential. Here, we introduce shinyDeepDR to bridge this gap by providing an intuitive and user-friendly web platform to access DeepDR. In the broader scope, we envision that tools like shinyDeepDR will advance cancer research by making sophisticated computational models more FAIR (findable, accessible, interoperable, and reusable).
Advancing precision oncology requires accurate prediction of treatment response and accessible prediction models. To this end, we present shinyDeepDR, a user-friendly implementation of our innovative deep learning model, DeepDR, for predicting anti-cancer drug sensitivity. The web tool makes DeepDR more accessible to researchers without extensive programming experience. Using shinyDeepDR, users can upload mutation and/or gene expression data from a cancer sample (cell line or tumor) and perform two main functions: “Find Drug,” which predicts the sample’s response to 265 approved and investigational anti-cancer compounds, and “Find Sample,” which searches for cell lines in the Cancer Cell Line Encyclopedia (CCLE) and tumors in The Cancer Genome Atlas (TCGA) with genomics profiles similar to those of the query sample to study potential effective treatments. shinyDeepDR provides an interactive interface to interpret prediction results and to investigate individual compounds. In conclusion, shinyDeepDR is an intuitive and free-to-use web tool for in silico anti-cancer drug screening.