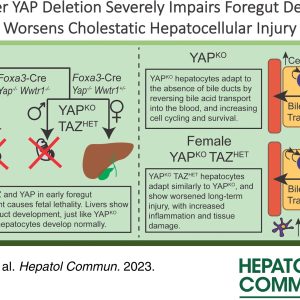
Dr. Mo Ebrahimkhani, Associate Professor of Pathology, and Dr. Ziv Bar-Joseph, Computational Biology at CMU, co-author an article published in Genome Biology, entitled, “TraSig: inferring cell-cell interactions from pseudotime ordering of scRNA-Seq data.”
Click here to read the full article
Li D, Velazquez JJ, Ding J, Hislop J, Ebrahimkhani MR, Bar-Joseph Z. TraSig: inferring cell-cell interactions from pseudotime ordering of scRNA-Seq data. Genome Biol. 2022 Mar 7;23(1):73. doi: 10.1186/s13059-022-02629-7. PMID: 35255944; PMCID: PMC8900372.
Abstract
A major advantage of single cell RNA-sequencing (scRNA-Seq) data is the ability to reconstruct continuous ordering and trajectories for cells. Here we present TraSig, a computational method for improving the inference of cell-cell interactions in scRNA-Seq studies that utilizes the dynamic information to identify significant ligand-receptor pairs with similar trajectories, which in turn are used to score interacting cell clusters. We applied TraSig to several scRNA-Seq datasets and obtained unique predictions that improve upon those identified by prior methods. Functional experiments validate the ability of TraSig to identify novel signaling interactions that impact vascular development in liver organoids.